3.05 with documenation
Showing
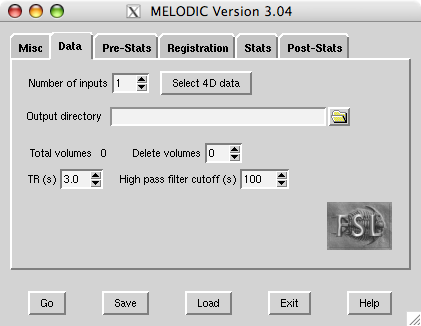
- doc/gui.png 0 additions, 0 deletionsdoc/gui.png
- doc/index.html 150 additions, 34 deletionsdoc/index.html
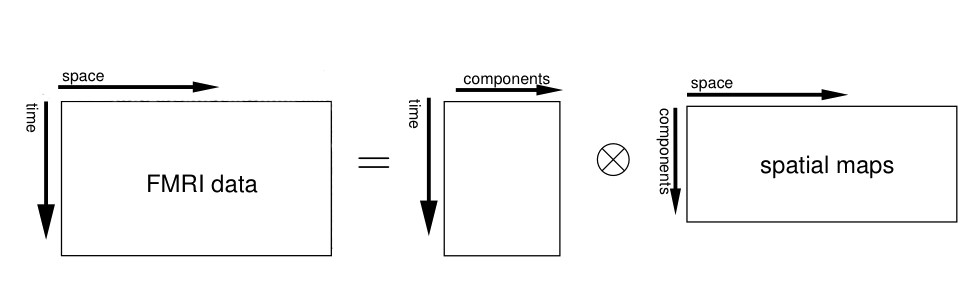
- doc/pica_diag.png 0 additions, 0 deletionsdoc/pica_diag.png
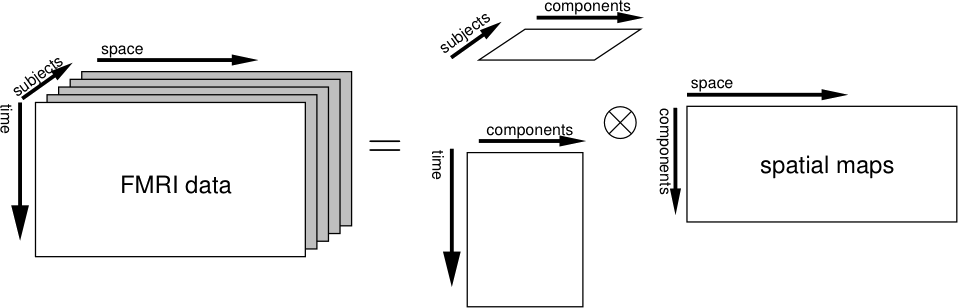
- doc/tica_diag.png 0 additions, 0 deletionsdoc/tica_diag.png
- melodic.h 1 addition, 1 deletionmelodic.h
- melreport.cc 5 additions, 5 deletionsmelreport.cc
doc/gui.png
0 → 100644
21.4 KiB
doc/pica_diag.png
0 → 100644
1.13 MiB
doc/tica_diag.png
0 → 100644
1.13 MiB